Metatranscriptomic analysis shows functional alterations in subgingival biofilm in young smokers with periodontitis: a pilot study
DOI:
https://doi.org/10.1590/1678-7757-2024-003Keywords:
Smoking, Periodontitis, RNA-Seq, Host-pathogen interactions, Gene expression, Oral microbiology, Non-invasive diagnosticsAbstract
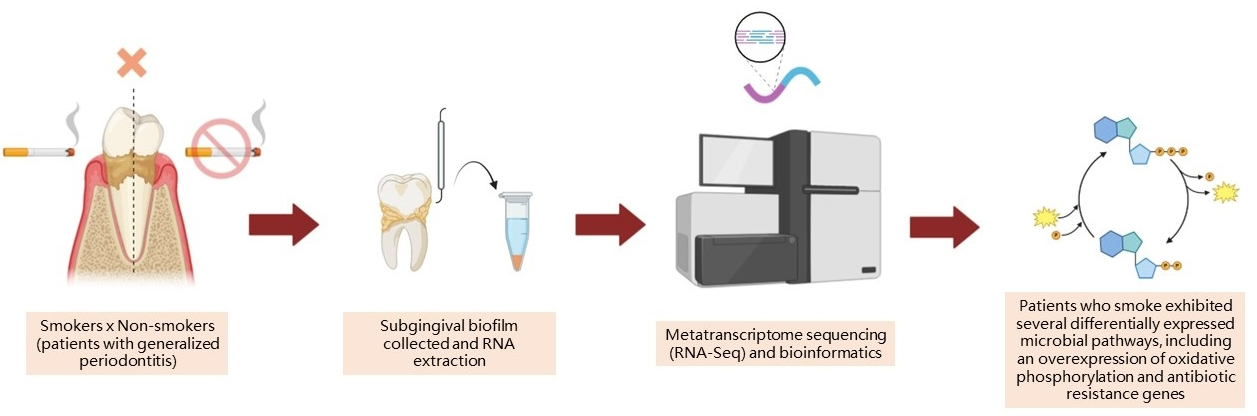
This study aimed to assess the influence of smoking on the subgingival metatranscriptomic profile of young patients affected by stage III/IV and generalized periodontal disease. Methodology: In total, six young patients, both smokers and non-smokers (n=3/group), who were affected by periodontitis were chosen. The STROBE (Strengthening the Reporting of Observational Studies in Epidemiology) guidelines for case-control reporting were followed. Periodontal clinical measurements and subgingival biofilm samples were collected. RNA was extracted from the biofilm and sequenced via Illumina HiSeq. Differential expression analysis used Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment, and differentially expressed genes were identified using the Sleuth package in R, with a statistical cutoff of ≤0.05. Results: This study found 3351 KEGGs in the subgingival biofilm of both groups. Smoking habits altered the functional behavior of subgingival biofilm, resulting in 304 differentially expressed KEGGs between groups. Moreover, seven pathways were modulated: glycan degradation, galactose metabolism, glycosaminoglycan degradation, oxidative phosphorylation, peptidoglycan biosynthesis, butanoate metabolism, and glycosphingolipid biosynthesis. Smoking also altered antibiotic resistance gene levels in subgingival biofilm by significantly overexpressing genes related to beta-lactamase, permeability, antibiotic efflux pumps, and antibiotic-resistant synthetases. Conclusion: Due to the limitations of a small sample size, our data suggest that smoking may influence the functional behavior of subgingival biofilm, modifying pathways that negatively impact the behavior of subgingival biofilm, which may lead to a more virulent community.
Downloads
References
Nociti FH, Casati MZ, Duarte PM. Current perspective of the impact of smoking on the progression and treatment of periodontitis. Periodontol2000. 2015;67(1):187-210. doi: 10.1111/prd.12063
Champagne BM, Sebrié EM, Schargrodsky H, Pramparo P, Boissonnet C, Wilson E. Tobacco smoking in seven Latin American cities: the CARMELA study. Tob Control. 2010;19(6):457-62. doi: 10.1136/tc.2009.031666
Graetz C, Sälzer S, Plaumann A, Schlattmann P, Kahl M, Springer C, et al. Tooth loss in generalized aggressive periodontitis: prognostic factors after 17 years of supportive periodontal treatment. J Clin Periodonto.l 2017;44(6):612–9. doi: 10.1111/jcpe.12725
Kanmaz B, Lappin DF, Nile CJ, Buduneli N. Effects of smoking on non‐surgical periodontal therapy in patients with periodontitis Stage III or IV, and Grade C. J Periodontol. 2019;91(4):442–53. doi: 10.1002/JPER.19-0141
Mason MR, Preshaw PM, Nagaraja HN, Dabdoub SM, Rahman A, Kumar PS. The subgingival microbiome of clinically healthy current and never smokers. ISME J. 2014;9(1):268-72. doi: 10.1038/ismej.2014.114
Shah SA, Ganesan SM, Varadharaj S, Dabdoub SM, Walters JD, Kumar PS. The making of a miscreant: tobacco smoke and the creation of pathogen-rich biofilms. NPJ Biofilms Microbiomes. 2017;3:26. doi: 10.1038/s41522-017-0033-2
Duran‐Pinedo AE. Metatranscriptomic analyses of the oral microbiome. Periodontol 2000. 2020;85(1):28-45. doi: 10.1111/prd.12350
Tonetti MS, Greenwell H, Kornman KS. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J Periodontol. 2018;89(1):S159-72. doi: 10.1002/JPER.18-0006
Tong J, Zhao W, Lv H, Li W, Chen Z, Zhang C. Transcriptomic profiling in human decidua of severe preeclampsia detected by RNA sequencing. J Cell Biochem. 2018;119(1):607-15. doi: 10.1002/jcb.26221
Gómez MA, Belew AT, Navas A, Rosales-Chilama M, Murillo J, Dillon LA, et al. Early leukocyte responses in ex-vivo models of healing and non-healing human Leishmania (Viannia) panamensis infections. Frontiers in Cellular and Infection Microbiology. 2021;11:687607. doi: 10.3389/fcimb.2021.687607
von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP; STROBE Initiative. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. J Clin Epidemiol. 2008;61(4):344-9. doi: 10.1016/j.jclinepi.2007.11.008
Miller PD Jr, McEntire ML, Marlow NM, Gellin RG. An evidenced-based scoring index to determine the periodontal prognosis on molars. J Periodontol. 2014;85(2):214-25. doi: 10.1902/jop.2013.120675
Ainamo J, Bay I. Problems and proposals for recording gingivitis and plaque. Int Dent J. 1975;25(4):229-35.
Mühlemann HR, Son S. Gingival sulcus bleeding: a leading symptom in initial gingivitis. Helv Odontol Acta. 1971;15(2):107-13.
Andrews S. FastQC: a quality control tool for high throughput sequence data [Software]. Cambridge: Babraham Bioinformatics; 2010. Available from: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114-20. doi: 10.1093/bioinformatics/btu170
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357-9. doi: 10.1038/nmeth.1923
Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28(24):3211-7. doi: 10.1093/bioinformatics/bts611
Escapa IF, Huang Y, Chen T, Lin M, Kokaras A, Dewhirst FE, et al. Construction of habitat-specific training sets to achieve species-level assignment in 16S rRNA gene datasets. Microbiome. 2020;8(1):65. doi: 10.1186/s40168-020-00841-w
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011 May 15;29(7):644-52. doi: 10.1038/nbt.1883
Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34(5):525–7. doi: 10.1038/nbt.3519
Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 2017;14(7):687-90. doi: 10.1038/nmeth.4324
Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015;44(D1):D457-62. doi: 10.1093/nar/gkv1070
Kim J, Kim MS, Koh AY, Xie Y, Zhan X. FMAP: Functional Mapping and Analysis Pipeline for metagenomics and metatranscriptomics studies. BMC Bioinformatics. 2016;17(1). doi: 10.1186/s12859-016-1278-0
Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45(D1):D566-73. doi: 10.1093/nar/gkw1004
Ching T, Huang S, Garmire LX. Power analysis and sample size estimation for RNA-Seq differential expression. RNA. 2014;20(11):1684-96. doi: 10.1261/rna.046011.114
Kulkarni R, Antala S, Wang A, Amaral FE, Rampersaud R, LaRussa SJ, et al. Cigarette smoke increases Staphylococcus aureus biofilm formation via oxidative stress. Infect Immun. 2012;80(11):3804-11.10.1128/IAI.00689-12
Jia YJ, Liao Y, He YQ, Zheng MQ, Tong XT, Xue WQ, et al. Association between oral microbiota and cigarette smoking in the chinese population. Front Cell Infect Microbiol. 2021;11:658203. doi: 10.3389/fcimb.2021.658203
Wu J, Peters BA, Dominianni C, Zhang Y, Pei Z, Yang L, et al. Cigarette smoking and the oral microbiome in a large study of American adults. The ISME Journal. 2016;10(10):2435–46. doi: 10.1038/ismej.2016.37
Schwarz MG, Antunes D, Corrêa PR, Silva-Gonçalves AJ, Malaga W, Caffarena ER, et al. Mycobacterium tuberculosis and M. bovis BCG
Moreau fumarate reductase operons produce different polypeptides that may be related to non-canonical functions. Front Microbiol. 2021;11:624121. doi: 10.3389/fmicb.2020.624121
Zhang L, Liu C, Jiang Q, Yin Y. Butyrate in energy metabolism: there is still more to learn. Trends Endocrinol Metab. 2021;32(3):159-69. doi: 10.1016/j.tem.2020.12.003
Perinbam K, Chacko JV, Kannan A, Digman MA, Siryaporn A. A shift in central metabolism accompanies virulence activation in Pseudomonas aeruginosa. mBio. 2020;11(2):e02730-18. doi: 10.1128/mBio.02730-18
Hreha TN, Foreman S, Duran-Pinedo AE, Morris AP, Manucha W, Jones JR, et al. The three NADH dehydrogenases of Pseudomonas aeruginosa: their roles in energy metabolism and links to virulence. PLoS One. 2021;16(2):e0244142–2. doi: 10.1371/journal.pone.0244142
Cogo K, Calvi BM, Mariano FS, Franco GC, Gonçalves RB, Groppo FC. The effects of nicotine and cotinine on Porphyromonas gingivalis colonisation of epithelial cells. Archives of Oral Biology. 2009;54(11):1061–7. doi: 10.1016/j.archoralbio.2009.08.001
Cogo K, Andrade A, Labate CA, Bergamaschi CC, Berto LA, Franco GC, et al. Proteomic analysis of Porphyromonas gingivalis exposed to nicotine and cotinine. J Periodontal Res. 2012;47(6):766-75. doi: 10.1111/j.1600-0765.2012.01494.x
Settem RP, Honma K, Nakajima T, Phansopa C, Roy S, Stafford GP, et al. A bacterial glycan core linked to surface (S)-layer proteins modulates host immunity through Th17 suppression. Mucosal Immunol. 2013;6(2):415-26. doi: 10.1038/mi.2012.85
Chiu CY, Chou HC, Chang LC, Fan WL, Dinh MCV, Kuo YL, Chung WH, Lai HC, Hsieh WP, Su SC. Integration of metagenomics-metabolomics reveals specific signatures and functions of airway microbiota in mite-sensitized childhood asthma. Allergy. 2020;75(11):2846-2857. doi: 10.1111/all.14438
Lynskey NN, Reglinski M, Calay D, Siggins MK, Mason JC, Botto M, et al. Multi-functional mechanisms of immune evasion by the streptococcal complement inhibitor C5a peptidase. PLoS Pathog. 2017;13(8):e1006493. doi: 10.1371/journal.ppat.1006493
Huang R, Li M, Ye M, Yang K, Xu X, Gregory RL. Effects of nicotine on Streptococcus gordonii growth, biofilm formation, and cell aggregation. Appl Environ Microbiol. 2014 Dec;80(23):7212-8. doi: 10.1128/AEM.02395-14
Silva RV, Rangel TP, Corrêa MG, Monteiro MF, Casati MZ, Ruiz KG, et al. Smoking negatively impacts the clinical, microbiological, and immunological treatment response of young adults with Grade C periodontitis. J Periodontal Res. 2022 Dec;57(6):1116-1126. doi: 10.1111/jre.13049
Altabtbaei K, Maney P, Ganesan SM, Dabdoub SM, Nagaraja HN, Kumar PS. Anna Karenina and the subgingival microbiome associated with periodontitis. Microbiome. 2021;9(1):97. doi: 10.1186/s40168-021-01056-3
Downloads
Published
Versions
- 2024-10-01 (2)
- 2024-08-16 (1)
Issue
Section
License
Copyright (c) 2024 Journal of Applied Oral Science

This work is licensed under a Creative Commons Attribution 4.0 International License.
Todo o conteúdo do periódico, exceto onde está identificado, está licenciado sob uma Licença Creative Commons do tipo atribuição CC-BY.

